Introduction
Clinical genetic testing is a powerful diagnostic tool for neurological disorders. The utility of genetic testing can be diminished by the large number of variants of uncertain significance (VUS). Variant classification for neurological disorders has additional challenges because clinical evidence is often limited. The biggest limitation on classification of potential splicing variants is that disease-relevant tissues are not readily accessible for neurological disorders. However, a recent oncology publication successfully showed whole blood could be used as the source of RNA.1
Methods
Recently, we undertook an initiative to measure the impact of targeted RNA analysis on patients undergoing genetic testing for a neurological indication at our laboratory. We had two specific objectives for this study: The first was to evaluate whether whole blood can be used for RNA studies for neurological disorders; the second was to determine whether targeted RNA analysis is helpful to reclassify variants in genes associated with neurological disorders.
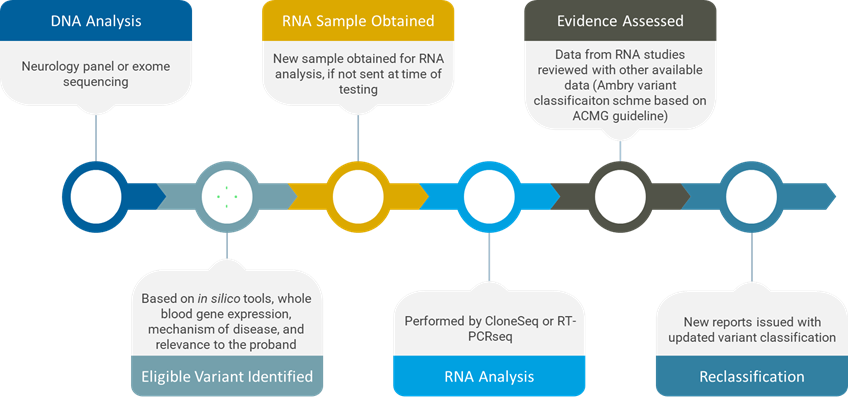
For this study, we selected 32 potential splicing variants detected in 28 patients. Three cases were from single gene testing, 14 from multi-gene panel, and 11 from exome sequencing.
Germline variants were considered for RNA studies based on the following eligibility criteria:
• In silico tools predict an impact on splicing
• Genes have sufficient expression in whole blood
• Mechanism of disease is consistent with splicing impact
• The results will be clinically relevant to the proband

For cases with variants meeting these criteria, RNA was extracted from whole blood. Our RNA analysis was primarily performed by either CloneSeq or RT-PCRseq, which provided both qualitative and quantitative data.
After RNA analysis, we reassessed variants with new RNA data, using the Ambry variant classification scheme, which is based on the 2015 ACMG/AMP guidelines.
Results
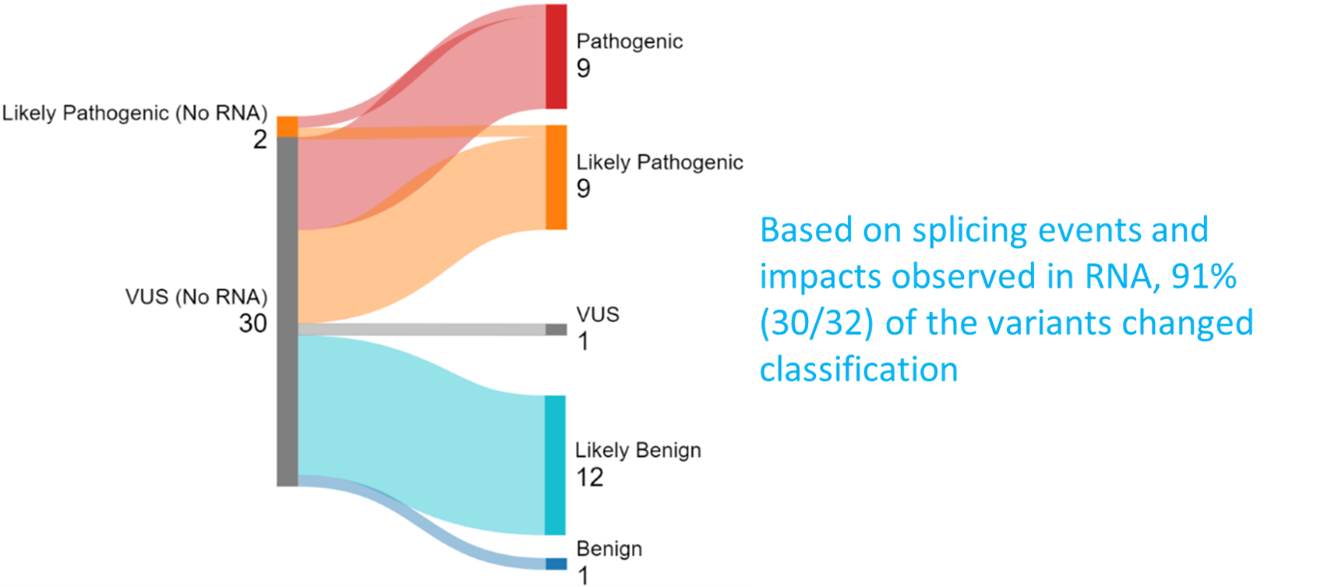
Overall, our RNA analysis impacted the classification of 30 out of 32 variants, or 93% of the variants assessed in this study. If a variant was upgraded or downgraded, a new report was issued to the patient.
• There were 30 VUSs. Most variants were either upgraded or downgraded, using RNA evidence.
o 16 variants or 53% of the variants were upgraded.
o 13 variants or 43% of the variants were downgraded.
• We evaluated 2 likely pathogenic variants. One of them was upgraded to pathogenic after RNA studies.
• Two variants remained the same classification after RNA analysis.
o One variant was kept at likely pathogenic due to atypical clinical presentation of the affected family. Another variant was missense. While we were able to rule out a splicing impact, it may still affect protein function and remained a VUS.
Case Study
As an example of the power of direct RNA evidence, we identified an intronic variant in FMR1 in a young male with a history of developmental delay, autistic spectrum disorder, hypotonia and other features. Both parents were unaffected. Fragile X was suspected, and the patient underwent FMR1 repeat analysis, which was negative. He also had a negative SNP array. He was then tested on our neurodevelopmental panel and found to carry a hemizygous intronic deletion in FMR1.
FMR1 is associated with Fragile X syndrome, which is the most common inherited form of developmental delay. In the vast majority of cases, it is caused by a trinucleotide repeat expansion in the 5’-untranslated region. This expansion essentially silences expression of the FMR1 gene, so it is loss-of-function.
Almost all patients with Fragile X syndrome have this trinucleotide repeat expansion, but a very small number of patients have been reported to have pathogenic mutations within the gene. Splicing mutations in the FMR1 gene are extremely rare.
The variant was de novo and absent in gnomAD. SpliceAI predicted that the variant disrupts the native donor site at 100% probability. Even with these lines of evidence, this variant remained a VUS, as we did not have direct RNA evidence. The patient then went on to get exome sequencing, with no additional reportable variants identified.
We offered RNA studies to this family and collected new blood samples from the patient and his mother to perform RNA analysis targeting this gene. The RNA studies provided strong evidence for pathogenicity, giving us sufficient evidence to upgrade this variant from VUS to pathogenic. A new report was issued to the clinician, providing a genetic diagnosis to the patient’s family.
Conclusion
Targeted RNA analysis using whole blood can provide useful information for variant classification in neurological disorders. In this cohort, RNA studies resulted in reclassification of 93% of the variants evaluated. RNA evidence improves the accuracy of genetic diagnosis of neurological disorders. This data demonstrates the impact of RNA data in reclassifying VUS and providing answers to patients and their families.
____
References and Notes:
1. https://pubmed.ncbi.nlm.nih.gov/37924330/



