
New discoveries are rapidly expanding our understanding of the human genome, and diagnostic laboratories use different approaches to interpret this knowledge. A challenge for laboratories is translating vast amounts of published evidence to determine the clinical validity of gene-disease relationships, which need to be integrated into a patient’s diagnostic exome sequencing (DES) results.
Ambry’s newly published system for scoring the clinical validity of gene-disease relationships improves patient care by solving the problem of translating this important evidence.1 In doing so, this system provides a blueprint for interpreting exome results and designing multi-gene panel tests (MGPTs).
Read the full research published in Human Mutation
Background
DES analyzes all genes in the genome and can identify an underlying diagnosis to adjust a patient’s medical management2,3, benefiting the healthcare system from patients to payors. However, new information about genetic diseases is always being published and it is not always clear how much evidence is “enough” to make a diagnosis. If labs report DES results based on inadequate data, an incorrect diagnosis can be made, leading to missed opportunities for treatment and/or incorrect medical management.
Knowledge of disease-associated (Mendelian) genes within the genome is increasing, with ~9,000 still awaiting characterization as of 2015.4 Thankfully, much work of the past decade has helped to standardize reporting criteria for genetic variants that are found5,6.
However, there is no published methodical approach to interpreting the clinical validity of a gene-disease association – crucial information when interpreting DES results. As such, each lab currently interprets clinical validity according to its own criteria and dramatically different conclusions may result, even from the same patient sample.
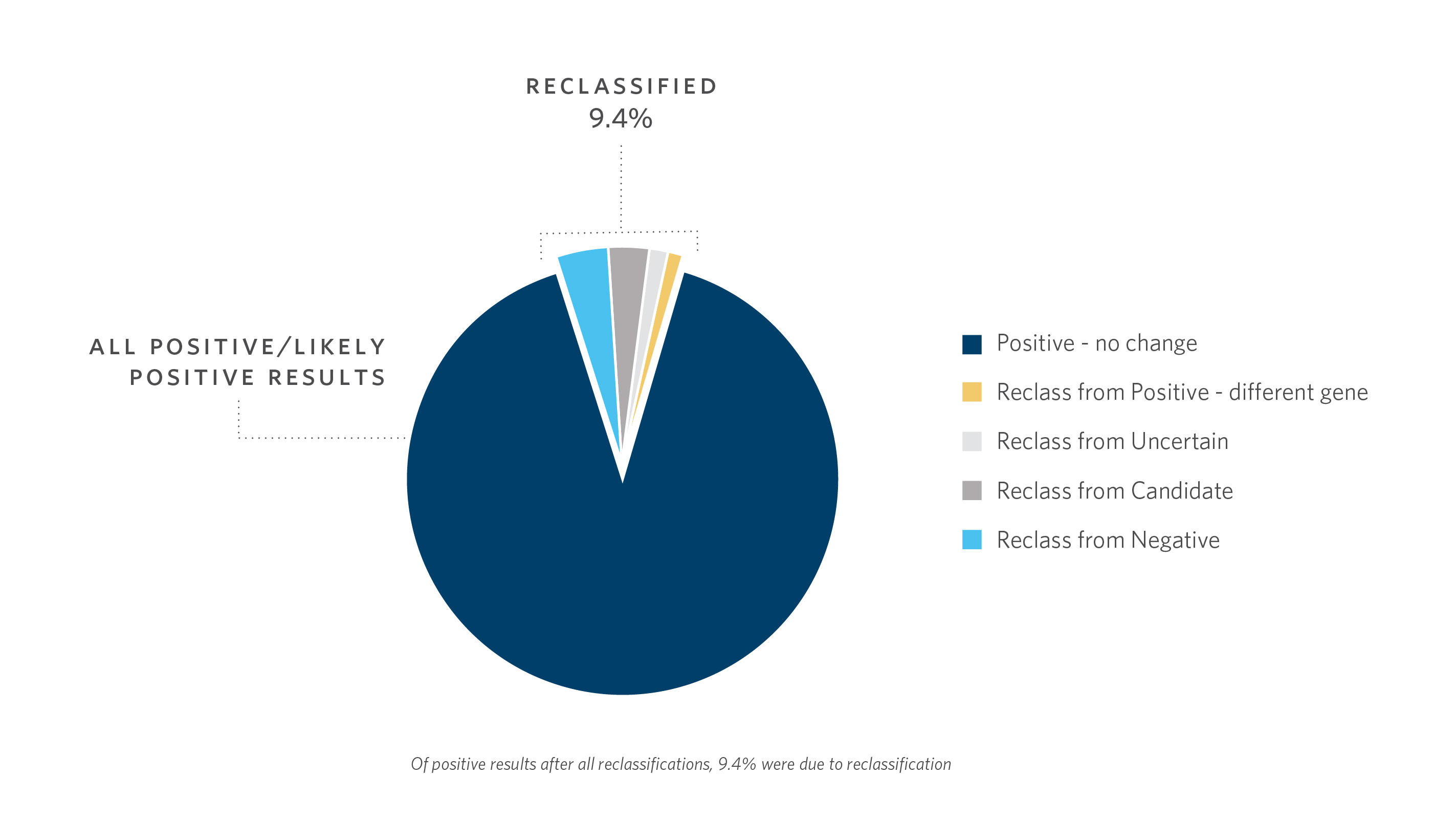
Reanalysis of previous cases by client requests or as part of updating a gene-disease database with new discoveries provided a diagnosis in 5% of previously unsolved cases, from our experience with the first >3,000 individual samples that underwent DES at Ambry.1
Ambry authors are giving a webinar presentation on their study and methodology- Register Here
Study in Brief
Ambry and Washington University researchers studied DES results on >3,000 independent patients who underwent the test at Ambry. Researchers combined personal communications with the curators of ClinGen7, existing guidelines for genetic variants8, and internal experience analyzing DES data since Ambry introduced the test to market in 2011.1,9
Key Results
- The scoring system assigned points for categories of evidence (listed below). Scores of 8+ were classified as “Characterized” genes.
- Number of unrelated patients (1-4 pts): Genetic “burden” in human patients
- Other statistical evidence (0-1 pt): Extensive correlation of genetic finding with disease
- Number of publications reporting independent patients (0-3 pts): Replication of findings
- Gene function (0-4 pts): Gene expression or function consistent with disease
- Gene disruption (0-2 pts): In vitro evidence that gene disruption leads to cellular pathology
- Model organism (0-2 pts): In vivo models from other organisms supporting a role of gene in disease
- Using the scoring system, reclassification of DES cases >1 year old contributed ~10% of the “Positive/Likely Positive” diagnostic yield in characterized genes
What This Means for Your Practice
- This is the first published scoring system to standardize clinical validity evaluation of gene-disease relationships, which helps clarify the meaning of patient results
- In the absence of industry-wide consensus about how often to reanalyze previously negative DES cases, this study makes suggestions so new diagnoses can be reported to a clinician
- Candidate genes should follow a fully vetted analysis and reporting process, such as that which Ambry researchers established and published8
- Public data sharing is imperative to help patients and families gain a diagnosis informed by rapidly evolving knowledge,10 particularly for DES
To learn more about ExomeNext, Ambry’s diagnostic exome sequencing, click here.
To read Ambry’s more about Ambry’s gene classification processes, click here.
References
1. Smith E, et al. Classification of genes: Standardized clinical validity assessment of gene-disease associations aids diagnostic exome analysis and reclassifications. Hum Mutat. 2017 May;38(5):600-08.
2. Soden SE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014 Dec 3;6(265):265ra168.
3. Srivastava S, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014 Oct;76(4):473-83.
4. Chong JX, et al. The genetic basis of Mendelian phenotypes: Discoveries, challenges, and opportunities. Am J Hum Genet. 2015 Aug 6;97(2):199-215.
5. Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405-24.
6. Amendola LM, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016 Jun 2;98(6):1067-76.
7. Clinical Genome Resource (ClinGen). Clinical Validity Curation Spreadsheet. [Internet]. Available at https://www.clinicalgenome.org/working-groups/gene-curation/projectsinitiatives/gene-disease-clinical-validity-scoring-matrix/. Accessed October 2016 and May 4, 2017.
8. MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014 Apr 24;508(7497):469-76.
9. Farwell K, et al. Candidate-gene criteria for clinical reporting: Diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med. 2017 Feb;19(2):224-235.
10. Harrison SM, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017 Mar 16. [Epub ahead of print.]



