In 2013, Emma’s parents1 hoped that a new genetic test, whole exome sequencing (WES), would reveal why their 2-year-old daughter developed epileptic encephalopathy. However, when Emma’s WES results were negative, her parents were disappointed. How could they examine the whole exome and not find an answer?
For patients with rare genetic disorders like Emma, the journey to diagnosis can be a long and difficult one. The diagnostic odyssey can take years of medical appointments, tests, and misdiagnoses before a patient finally receives an accurate diagnosis. For those with epileptic encephalopathy, this journey can be particularly challenging. However, advances in our understanding of gene-disease relationships have made it possible to end the diagnostic odyssey for some patients like Emma.
Epileptic encephalopathy is a rare neurological disorder that is characterized by seizures and intellectual disability. It is often caused by genetic mutations that affect the brain's ability to function properly. Because there are many different genes that can cause epileptic encephalopathy, whole exome sequencing (WES) is utilized by many healthcare providers to determine the underlying cause.
A year after Emma’s initial WES test, her geneticist requested a reanalysis of Emma’s WES data. Many new gene-disease associations had been established for epileptic encephalopathy and numerous other associations were emerging. While no definitive diagnosis was made, an uncertain alteration in an emerging gene-disease association was noted. Within the year of Emma’s WES report, two case studies were published describing de novo KCNH5 variants in patients with epileptic encephalopathy2, 3. The gene-disease validity for this association was characterized as uncertain so no definitive diagnosis could be made. However, Emma’s family was matched with researchers interested in further studying the association.
Gene-disease associations are a crucial aspect of modern medicine. They help us understand the genetic basis of diseases and develop targeted treatments. Gene-disease validity refers to the strength of evidence supporting a genetic association with a particular disease. It is determined by evaluating the available scientific evidence, including genetic and clinical data, to assess the likelihood that a particular gene or genetic variant contributes to the development or risk of a specific disease. The higher the gene-disease validity, the stronger the evidence supporting the association between the gene and disease. The level of validity can range from strong, indicating a high likelihood of a causal relationship, to limited indicating a need for further investigation.4, 5, 6
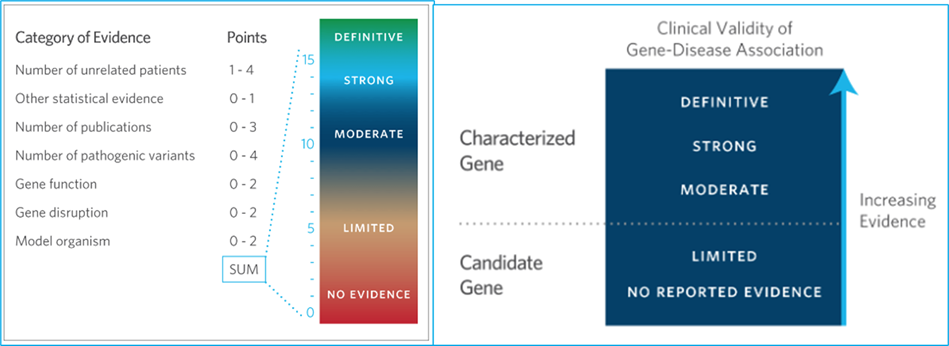
Classification of Genes: Standardized Clinical Validity Assessment of Gene-Disease Associations Aids Diagnostic Exome Analysis and Reclassifications PMID: 28106320
Nine years after her initial WES, Emma’s parents received the phone call they had been waiting for: “We have a diagnosis.” In 2023, additional publications provided the necessary levels of evidence to characterize the gene-disease association as strong.7, 8, 9 KCNH5-related neurodevelopmental disorder is an autosomal dominant disorder that typically occurs de novo. The phenotype includes infantile-onset of multiple seizure types, developmental delay, intellectual disability, and typically normal brain MRI. Severity can be variable and may correlate to genotype.
In the 9 years since her initial testing, Emma’s geneticist retired, and their genetic counselor moved away. They had lost the close contact they once had with the office. Ambry’s Patient for Life Program ensured that Emma’s diagnosis was not lost to follow-up. As new gene-disease relationships are established and the phenotypic spectrum of relationships emerge, Ambry continually reanalyzes all exome cases and proactively contacts healthcare providers when their patients are impacted.
Ending the diagnostic odyssey for patients with rare genetic disorders like epileptic encephalopathy is an important goal for doctors and researchers. Exome sequencing has made it possible to identify new gene-disease relationships and provide accurate diagnoses for patients who have been searching for answers for years.
Notes:
1. Names and patient details were changes to protect the identify of the patient and family.
2. Doi:10.1111/epi.1221
3. Doi:10.1523/JNEUROSI.2307-13.2013
4. Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100(6):895-906. doi:10.1016/j.ajhg.2017.04.015
5. Smith ED, Radtke K, Rossi M, et al. Classification of Genes: Standardized Clinical Validity Assessment of Gene-Disease Associations Aids Diagnostic Exome Analysis and Reclassifications. Hum Mutat. 2017;38(5):600-608. doi:10.1002/humu.23183
6. ClinGen: https://www.clinicalgenome.org/curation-activities/gene-disease-validity/
7. Doi:10.1212/WLN.0000000000201492
8.Doi:10.3389/fped.2022.858008
9. Doi:10.1186/s13023-022-02499-z




