One in every eight women will develop breast cancer in her lifetime.1 Considering there are 167.5 million women currently living in the United States,2 on a population level, that is a large number of women who will develop breast cancer. The vast majority of breast cancer cases are due to a combination of factors including, but not limited to: environment/exposures, lifestyle, and a woman’s age. These are considered sporadic cases, and they make up about 85% of all breast cancer cases. A smaller percentage of breast cancer cases—10-15%—are due to hereditary factors, or harmful differences in genes that a person is born with and might increase the risk for cancer. Of these 10-15% of cases, only about a quarter are due to a known genetic cause. That means that most cases of breast cancer that appear to run in a family still do not have an identifiable heritable cause.3
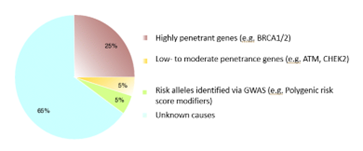
This “missing heritability” understandably has created a huge drive in the scientific community to discover yet-unknown genes that may be linked with an increased risk of breast cancer. We have many genes that play roles in protecting us from cancer, and harmful differences in these genes can therefore lower our defenses and increase cancer risk. But how are new breast cancer-linked genes identified? What kind of information is necessary to prove that harmful differences in a specific gene are responsible for causing breast cancer? This is where gene-disease validity (GDV) assessment comes in.
What is gene-disease validity? GDV tells us how certain we are that harmful differences in a particular gene cause a particular disease. It involves gathering evidence (clinical and experimental) and measuring the strength of that evidence toward or against a gene-disease relationship (GDR). When assessing a GDR for a very common disease like breast cancer, multiple confounding factors must be considered. This is because statistically speaking, the more people who have a disease of interest, the easier it is to identify something (a genetic difference or variant, for example) in that population.
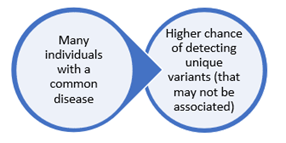
To explain this a bit more, here is a hypothetical example. Let’s say you have a group of 200 women in a population with what looks like familial or hereditary breast cancer. They are either diagnosed at a young age, or maybe other women in their family also have breast cancer. In this group, you want to look at possibly harmful variants in a newly implicated gene—we’ll call this Gene X—to see how common it is.
Then you take a group of 200 more women from the same population with no cancer, and you look for possible harmful variants in Gene X in them. You notice that 3% of women in the breast cancer group have variants in Gene X, and only 0.5% of women in the non-cancer group have variants in Gene X. Has a new breast cancer gene association been discovered? Not quite. While this bit of information may seem promising, much more data is needed to prove an association.
Here’s another simple example to explain: If I flip a coin only 4 times, and 1 of 4 times it lands on heads, then do I conclude that a coin toss yields a 25% chance of being heads and a 75% chance of being tails? We all know that statistically, the chances of heads or tails is 50%; if the coin toss experiment is repeated many more times, then the percentage will eventually level out to 50%. The same is true for GDV for breast cancer-linked genes. The clinical data needs to show replication over time, across several different studies, and in very large and diverse populations; this is because breast cancer has many different causes (genetic and non-genetic) and because it is so common; replication and large numbers help dilute any other effects that could bias results.
If initial, small studies are given too much weight in GDV assessment, then an association may be prematurely established. This can have the unfortunate consequence of a gene initially being reported as definitively linked with breast cancer, when after a few more studies emerge, new data shows that the gene is not at all associated with breast cancer and refutes that initial relationship. In addition to substantial clinical data, there must be sufficient evidence of a gene’s role in disease – what is the gene’s biological role? Is there evidence that harmful differences or mutations in a gene could contribute to tumor formation? This functional or experimental evidence must always complement the clinical data to complete the full GDV picture.
In short, establishing GDV for breast cancer-associated genes requires a lot more and/or many different lines of evidence to prove an association when compared to rare diseases. To learn more, check out this webinar on gene-disease validity assessment for breast cancer-associated genes.
___
1. http://www.cancer.gov/
2. https://www.census.gov/quickfacts/fact/table/US/LFE046222
3. Burwinkel, B., Rongxi, Y. (2011). Breast Cancer Familial Risk. In: Schwab, M. (eds) Encyclopedia of Cancer. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-16483-5_6695



