The ability of exome sequencing (ES) to detect variants across the genetic code makes it a powerful diagnostic tool, reducing the number of tests and time to diagnose patients with rare disorders. However, with this broad detection range comes the challenge of identifying which of hundreds or thousands of rare variants may be clinically meaningful for a patient. For this reason, ES is best performed in the setting of a parental trio—where samples from both biological parents are submitted at the time of testing alongside the patient’s sample. Having full ES data from both parents provides real-time inheritance information, narrowing the list of candidate variants for a patient. Parental trio ES analysis has been shown to reduce the number of variants of uncertain significance (VUS) and increase diagnostic rates. Nevertheless, there are limitations to submitting parental trios at the time of testing for various reasons, including known non-biological relationships such as adoption or use of gamete donors, and logistical considerations, such as parental estrangement and the absence of a parent at the appointment.
In this study, we investigated how different types of familial sample configurations (or FSC) impact the clinical utility and reclassification process of exome sequencing. We retrospectively reviewed nearly 11,000 exome sequencing cases tested over a 10-year period. Cases were categorized as parental trios, nonparental trios (three familial samples submitted, but representing one or fewer biological parents), duos and singletons (proband-only). To assess the clinical utility of testing under these FSCs, we looked at both the original testing outcome and the current reported outcome to account for any reclassifications through the end of 2021. We also looked at the sources of evidence used for reclassification.
We found that about 70% of cases submitted were parental trios, 13% percent were singletons, 11% were duos and 5% were nonparental trios. Unsurprisingly, we found that parental trios exhibited a significantly higher diagnostic rate and lower VUS rates compared to other FSCs.
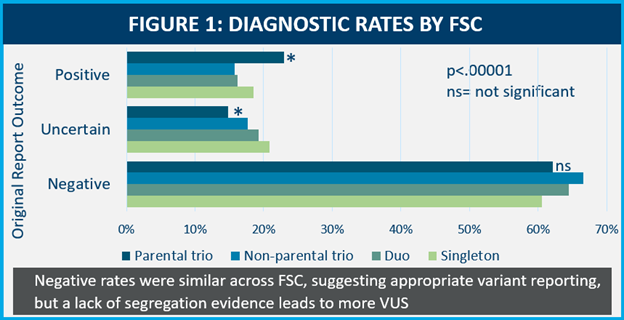
Notably, negative rates were similar across all FSCs, suggesting that appropriate variant detection and reporting is occurring, but without both biological parental samples, there is a lack of available segregation evidence (a way to determine if a variant was inherited or de novo in the patient), leading to higher VUS rates.
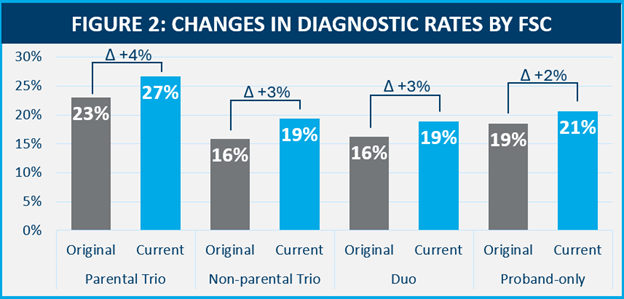
Evidence-driven reclassification increased diagnostic yield for all FSCs, but parental trios saw the largest diagnostic gain.
Evidence used for reclassification came from 37 different categories. New published evidence, often leading to the characterization of new gene disease relationships (GDR), was found to be the primary driver for proactive reclassifications across all FSCs, but the percentage of contribution for this line of evidence decreased in a correlative fashion with the number of samples provided. There were differences between the rates of other evidence used by FSC as well.
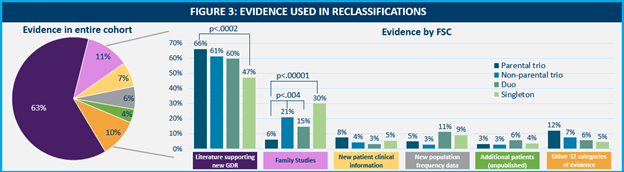
The observation that parental trios see the most considerable reclassification benefit, even when diverse lines of evidence are applied to FSC, highlights the importance of inheritance information for variant classification. The ability to know a variant is de novo (newly arising in a patient) may be the difference between a VUS and a diagnosis, and this information cannot be gathered in other ways. Therefore, even with our ever-expanding knowledge of GDRs and the ability to proactively integrate this new data into exome reclassification reports, FSCs without both biological parental samples face inherent limitations. These challenges can be compounded with other factors, such as underrepresentation in population databases, which may restrict the availability of additional lines of evidence. It is imperative that laboratories implement comprehensive methods for gathering diverse lines of evidence as they emerge, ensuring that every patient and variant is evaluated using the fullest extent of available data.
In addition, these findings underscore the necessity of ongoing, dynamic, evidence-driven reanalysis of exome sequencing data, like that offered in Ambry’s Patient for Life program. When approximately 30% of cases in the cohort are submitted for testing without a parental trio, this practice is crucial for maximizing the clinical utility of all FSCs and bridging the diagnostic gap between parental trios and other FSCs.



